Print a list of plots
print_plots.RdThis function iterates through a list of plots and prints them (without printing the list index).
Arguments
- plots
A list of
ggplot2::ggplot()objects.
Examples
gen_key <- read_key_csv(mctr_ex("example-gen-key.csv"))
#> Rows: 12 Columns: 7
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (2): Sex, Genotype
#> dbl (5): AS, SampNo, Spine, Met, Dia
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
gen_cort <- read_cortical_csv(mctr_ex("example-twice1.csv"),
mctr_ex("example-twice2.csv"),
gen_key)
#> Rows: 36 Columns: 75
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (44): SampName, MeasDate, ListDate, Filename, Site, Energy-I-Code, Contr...
#> dbl (26): SampNo, MeasNo, Integr.Time, Sigma, Support, Threshold, Unit, Data...
#> lgl (5): S-DOB, S-Remark, Meas-Rmk, RAW-Label, IMA-Label
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> Rows: 36 Columns: 75
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (43): SampName, MeasDate, ListDate, Filename, Site, Energy-I-Code, Contr...
#> dbl (27): SampNo, MeasNo, Integr.Time, Sigma, Support, Threshold, Unit, Data...
#> lgl (5): S-DOB, S-Remark, Meas-Rmk, RAW-Label, IMA-Label
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
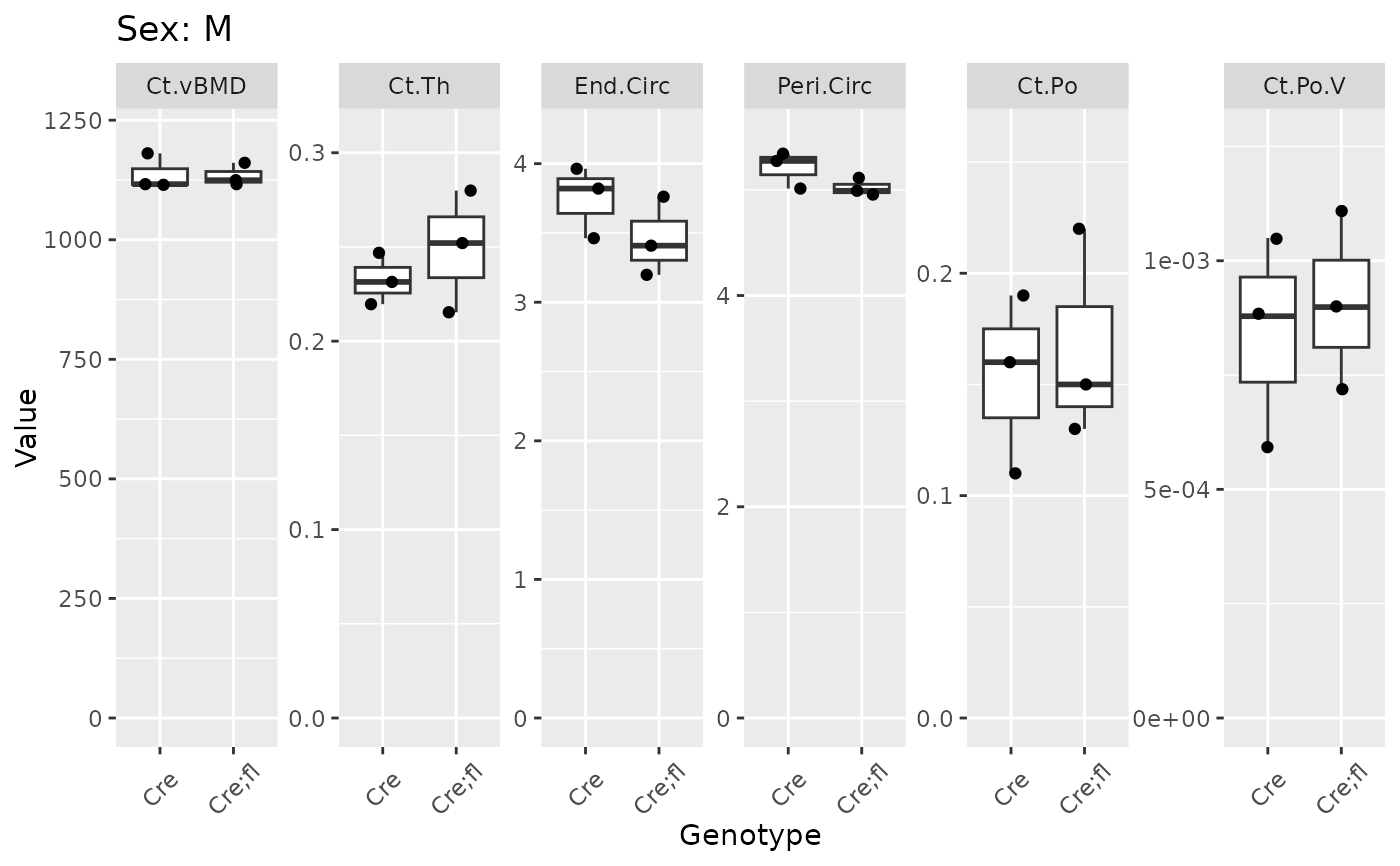
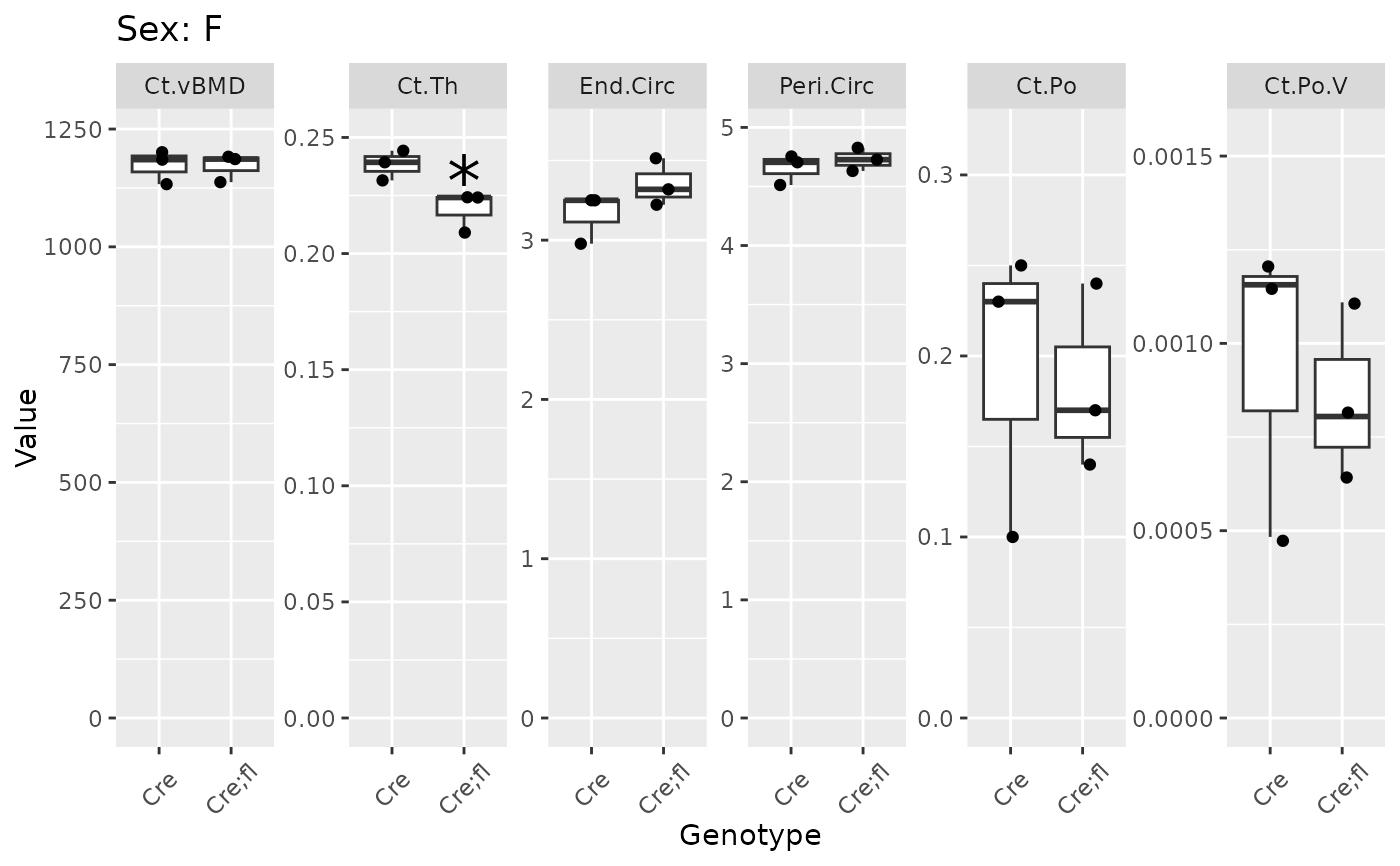
plot_groups(gen_cort |> dplyr::filter(Site == "Dia")) |> print_plots()